Sensitivity Analysis¶
Sensitivity Base class¶
-
class
pystran.SensitivityAnalysis(parsin)[source]¶ Base class for the Sensitivity Analysis
Parameters: parsin : list
ModPar class instances in list or list of (min, max,’name’)-tuples
Interaction with models¶
Morris Screening method¶
Main Morris screening methods¶
-
class
pystran.MorrisScreening(parsin, ModelType='external')[source]¶ Morris screening method, with the improved sampling strategy, selecting a subset of the trajectories to improve the sampled space. Working with groups is possible.
Parameters: parsin : list
either a list of (min,max,’name’) values, [(min,max,’name’),(min,max,’name’),...(min,max,’name’)] or a list of ModPar instances
ModelType : pyFUSE | PCRaster | external
Give the type of model working withµ
Notes
- Original Matlab code from:
- http://sensitivity-analysis.jrc.it/software/index.htm
Original method described in [M1], improved by the optimization of [M2]. The option to work with groups is added, as described in [M2].
References
[M1] Morris, Max D. Factorial Sampling Plans for Preliminary Computational Experiments. Technometrics 33, no. 2 (1991): 161–174. [M2] (1, 2, 3) Campolongo, Francesca, Jessica Cariboni, and Andrea Saltelli. An Effective Screening Design for Sensitivity Analysis of Large Models. Environmental Modelling & Software 22, no. 10 (October 2007): 1509–1518. http://linkinghub.elsevier.com/retrieve/pii/S1364815206002805. [M3] (1, 2) Saltelli, Andrea, Marco Ratto, Terry Andres, Francesca Campolongo, Jessica Cariboni, Debora Gatelli, Michaela Saisana, and Stefano Tarantola. Global Sensitivity Analysis, The Primer. John Wiley & Sons Ltd, 2008. Examples
>>> Xi = [(0.0,5.0,r'$X_1$'),(4.0,7.0,r'$X_2$'),(0.0,1.0,r'$X_3$'), (0.0,1.0,r'$X_4$'), (0.0,1.0,r'$X_5$'),(0.5,0.9,r'$X_6$')] >>> # Set up the morris class instance with uncertain factors Xi >>> sm = MorrisScreening(Xi,ModelType = 'external') >>> # calculate an optimized set of parameter sets to run model >>> OptMatrix, OptOutVec = sm.Optimized_Groups(nbaseruns=100, intervals = 4, noptimized=4, Delta = 0.4) >>> # Check the quality of the selected trajects >>> sm.Optimized_diagnostic(width=0.15) >>> #RUN A MODEL AND GET OUTPUT (EXTERNAL) -> get output >>> #Calculate the Morris screening diagnostics >>> sm.Morris_Measure_Groups(output) >>> #plot a barplot of mu, mustar and sigma (edgecolor and facecolor grey) >>> sm.plotmu(ec='grey',fc='grey') >>> sm.plotmustar(outputid = 1,ec='grey',fc='grey') >>> sm.plotsigma(ec='grey',fc='grey') >>> #plot the mu* sigma plain >>> sm.plotmustarsigma(zoomperc = 0.05, outputid = 1, loc = 2) >>> #export the results in txt file >>> sm.txtresults(name='MorrisTestOut.txt') >>> #export the results in tex-table >>> sm.latexresults(name='MorrisTestOut.tex')
Attributes
_ndim ( int) number of factors examined. In case the groups are chosen the number of factors is stores in NumFact and sizea becomes the number of created groups, (k) NumFact (int) number of factors examined in the case when groups are chosen intervals(p) (int) number of intervals considered in (0, 1) UB (ndarray) Upper Bound for each factor in list or array, (sizea,1) LB (ndarray) Lower Bound for each factor in list or array, (sizea,1) GroupNumber (int) Number of groups (eventually 0) GroupMat (ndarray) Array which describes the chosen groups. Each column represents a group and its elements are set to 1 in correspondence of the factors that belong to the fixed group. All the other elements are zero, (NumFact,GroupNumber) Delta (float) jump value to calculate screening intervals (int) number of intervals used in the sampling noptimized (int) r-value of the number of base runs are done in the optimize sampling OutMatrix (ndarray) not-optimized sample matrix OutFact (ndarray) not-optimzed matrix of changing factors Groupnumber (int) number of groups used sizeb (int) when using groups, sizeb is determined by the number of groups, otherwise the number of factors OptMatrix_b (ndarray) the not-adapted version of the OptMatrix, with all sampled values between, 0 and 1 parset2run (ndarrar) every row is a parameter set to run the model for. All sensitivity methods have this attribute to interact with base-class running -
Morris_Measure_Groups(Output)[source]¶ Calculates the Morris measures mu, mustar and sigma, the calculations with groups are in beta-version!
Can be used for multiple outputs
Parameters: Output : ndarray
if multiple outputs, every output in different column; the length of the outputs is the same as the optmatrix sampled
Returns: SAmeas : ndarray (_ndim*number of outputs, noptimized)
matrix with the elemenary effects, the factors in the rows, with different outputs after eachother; the columns take the repititions
OutMatrix : ndarray
Matrix of the output(s) values in correspondence of each point of each trajectory. For every output column, the factors are calculated and [Mu*, Mu, StDev] are put in the row When using groups, only Mu* for every group is given
Notes
The algorithm uses the self.OptOutMatrix and self.OptOutFact as the input calculations, but these can be given other input combinations too as long as it follows the Morris-method
-
Optimized_Groups(nbaseruns=500, intervals=4, noptimized=10, GroupMat=array([], dtype=float64), Delta='default')[source]¶ Optimization in the choice of trajectories for the Morris experiment. Starting from an initial set of nbaseruns, a set of noptimized runs is selected to use for the screening techique
Groups can be used to evaluate parameters together
Parameters: nbaseruns : int (default 500)
Total number of trajectories
intervals : int (default 4)
Number of levels
noptimized : int (default 10)
Final number of optimal trajectories
GroupMat : [NumFact,NumGroups]
Matrix describing the groups. Each column represents a group and its elements are set to 1 in correspondence of the factors that belong to the fixed group. All the other elements are zero.
Delta : ‘default’|float (0-1)
When default, the value is calculated from the p value (intervals), otherwise the given number is taken
Returns: OptMatrix/ self.OptOutMatrix : ndarray
Optimized sampled values giving the matrix too run the model for
OptOutVec/ self.OptOutFact : ndarray
Optimized sampled values giving the matrix indicating the factor changed at a specific line
Notes
The combination of Delta and intervals is important to get an good overview. The user is directed to [M3]
-
Sampling_Function_2(nbaseruns, LB, UB)[source]¶ Python version of the Morris sampling function
Parameters: nbaseruns : int
sample size
Returns: OutMatrix(sizeb*r, sizea) :
for the entire sample size computed In(i,j) matrices, values to run model for
OutFact(sizea*r,1) :
for the entire sample size computed Fact(i,1) vectors, indicates the factor changing at specific line
Notes
B0 is constructed as in Morris design when groups are not considered. When groups are considered the routine follows the following steps
1. Creation of P0 and DD0 matrices defined in Morris for the groups. This means that the dimensions of these 2 matrices are (GroupNumber,GroupNumber).
2. Creation of AuxMat matrix with (GroupNumber+1,GroupNumber) elements.
3. Definition of GroupB0 starting from AuxMat, GroupMat and P0.
4. The final B0 for groups is obtained as [ones(sizeb,1)*x0’ + GroupB0]. The P0 permutation is present in GroupB0 and it’s not necessary to permute the matrix (ones(sizeb,1)*x0’) because it’s already randomly created.
Adapted from the matlab version of 15 November 2005 by J.Cariboni
References
[M4] A. Saltelli, K. Chan, E.M. Scott, Sensitivity Analysis on page 68 ss [M5] - Campolongo, J. Cariboni, JRC - IPSC Ispra, Varese, IT
-
runTestModel(ai)[source]¶ Run a testmodel
Use a testmodel to get familiar with the method and try things out. The implemented model is the G Sobol function: testfunction with analytical solution, moire information, see [M3]
Parameters: ai : list
list with the input factors (equal size as number of factors)
Examples
>>> ai=[78, 12, 0.5, 2, 97, 33] >>> Xi = [(0.0,1.0,r'$X_1$'),(0.0,1.0,r'$X_2$'),(0.0,1.0,r'$X_3$'), (0.0,1.0,r'$X_4$'),(0.0,1.0,r'$X_5$'),(0.0,1.0,r'$X_6$')] >>> #set up the morris screening class >>> sm = MorrisScreening(Xi,ModelType = 'testmodel') >>> #Get an optimized set of trajectories >>> OptMatrix, OptOutVec = sm.Optimized_Groups(nbaseruns=100, intervals = 4, noptimized=4, Delta = 0.4) >>> #compare the selected trajects with the general >>> sm.Optimized_diagnostic(width=0.15) >>> #run a testmodel and get the outputs >>> sm.runTestModel(ai)
Testing the selected traject optimization¶
-
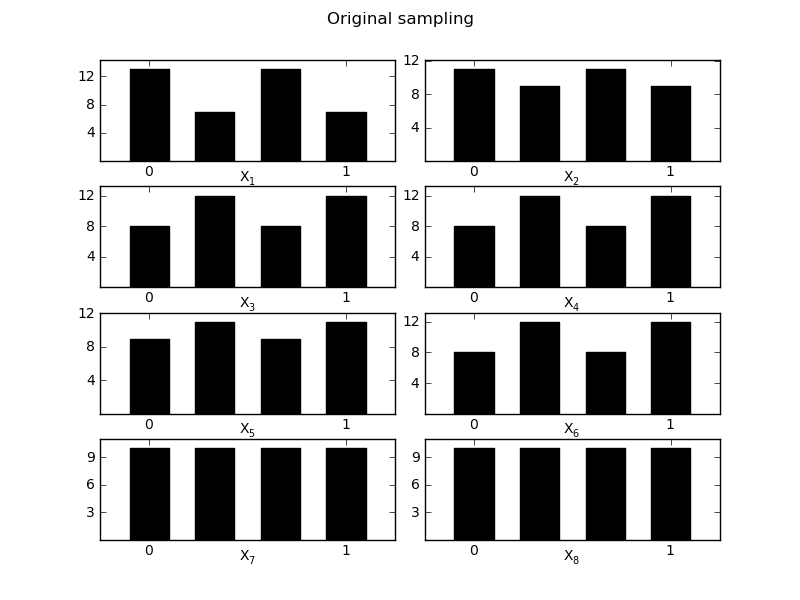
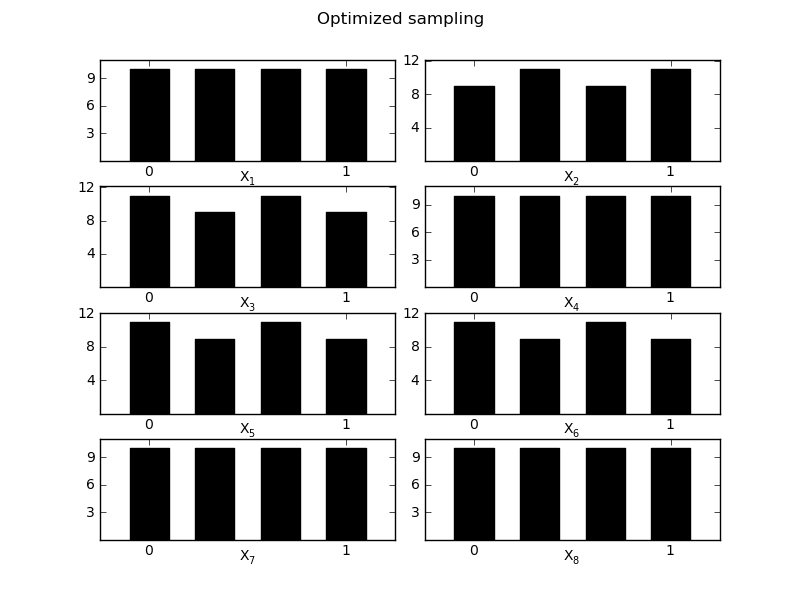
MorrisScreening.Optimized_diagnostic(width=0.1)[source]¶ Evaluate the optimized trajects in their space distirbution, evaluation is done based on the [0-1] boundaries of the sampling
Returns quality measure and 2 figures to compare the optimized version
Parameters: width : float
width of the bars in the plot (default 0.1)
Examples
>>> sm.Optimized_diagnostic() The quality of the sampling strategy changed from 0.76 with the old strategy to 0.88 for the optimized strategy
Output plots and exports¶
-
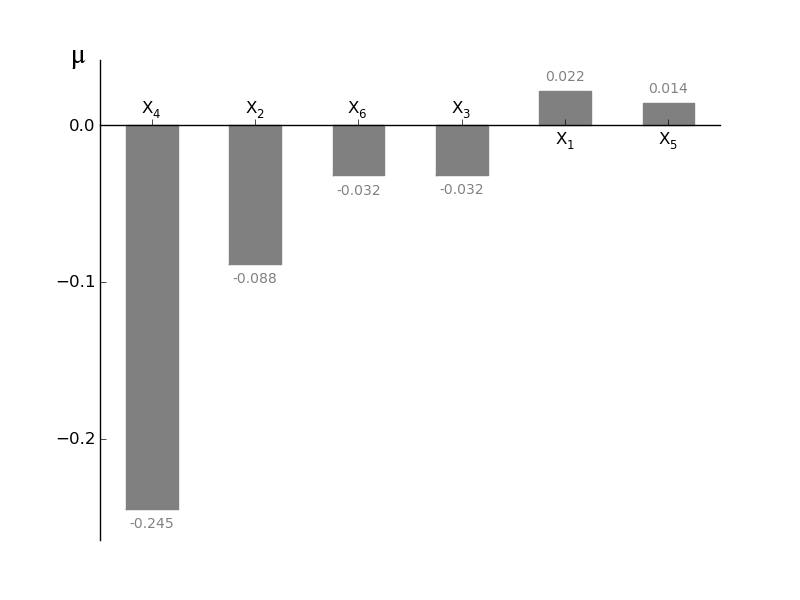
MorrisScreening.plotmu(width=0.5, addval=True, sortit=True, outputid=0, *args, **kwargs)[source]¶ Plot a barchart of the given mu
mu is a measure for the first-order effect on the model output. However the use of mu can be tricky because if the model is non-monotonic negative elements can be in the parameter distribution and by taking the mean of the variance (= mu!) the netto effect is cancelled out! Should always be used together with plotsigma in order to see whether higher order effects are occuring, high sigma values with low mu values can be caused by non-monotonicity of functions. In order to check this use plotmustar and/or plotmustarsigma
Parameters: width : float (0-1)
width of the bars in the barchart
addval : bool
if True, the morris mu values are added to the graph
sortit : bool
if True, larger values (in absolute value) are plotted closest to the y-axis
outputid : int
the output to use whe multiple are compared; starts with 0
*args, **kwargs : args
passed to the matplotlib.bar
Returns: fig : matplotlib.figure.Figure object
figure containing the output
ax1 : axes.AxesSubplot object
the subplot
-
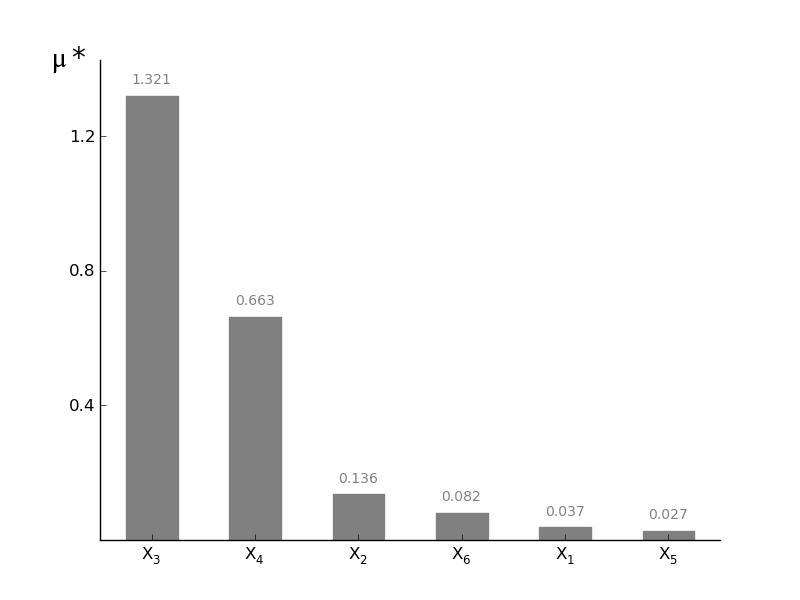
MorrisScreening.plotmustar(width=0.5, addval=True, sortit=True, outputid=0, *args, **kwargs)[source]¶ Plot a barchart of the given mustar
mu* is a measure for the first-order effect on the model output. By taking the average of the absolute values of the parameter distribution, the absolute effect on the output can be calculated. This is very useful when you are working with non-monotonic functions. The drawback is that you lose information about the direction of influence (is this factor influencing the output in a positive or negative way?). For the direction of influence use plotmustar!
Parameters: width : float (0-1)
width of the bars in the barchart
addval : bool
if True, the morris values are added to the graph
sortit : bool
if True, larger values (in absolute value) are plotted closest to the y-axis
outputid : int
the output to use whe multiple are compared; starts with 0
*args, **kwargs : args
passed to the matplotlib.bar; width is already used
Returns: fig: matplotlib.figure.Figure object
figure containing the output
ax1: axes.AxesSubplot object
the subplot
-
MorrisScreening.plotsigma(width=0.5, addval=True, sortit=True, outputid=0, *args, **kwargs)[source]¶ Plot a barchart of the given sigma
sigma is a measure for the higher order effects (e.g. curvatures and interactions). High sigma values typically occurs when for example parameters are correlated, the model is non linear,... The sum of all these effects is sigma. For linear models without any correlation sigma should be approximately zero.
Parameters: width : float (0-1)
width of the bars in the barchart
addval : bool
if True, the morris values are added to the graph
sortit : bool
if True, larger values (in absolute value) are plotted closest to the y-axis
outputid : int
the output to use whe multiple are compared; starts with 0
*args, **kwargs : args
passed to the matplotlib.bar
Returns: fig : matplotlib.figure.Figure object
figure containing the output
ax1 : axes.AxesSubplot object
the subplot
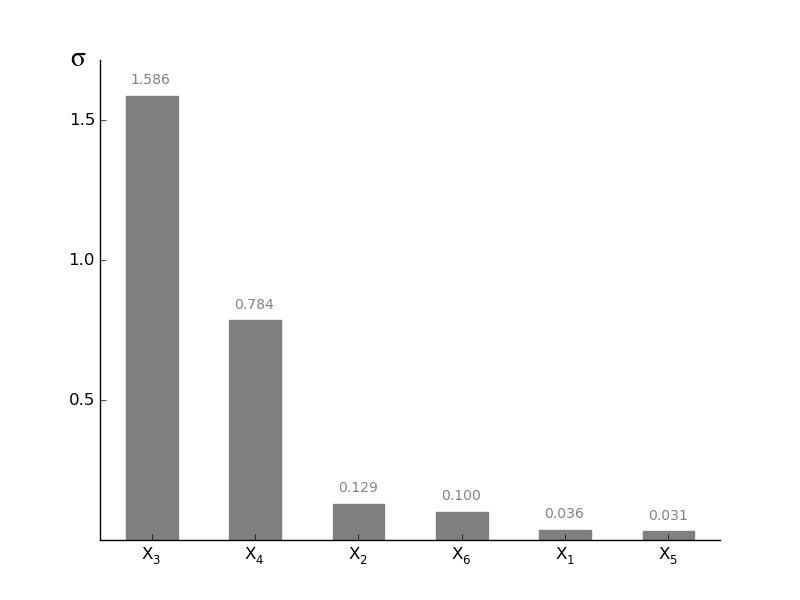
-
MorrisScreening.plotmustarsigma(outputid=0, zoomperc=0.05, loc=2)[source]¶ Plot the mu* vs sigma chart to interpret the combined effect of both.
Parameters: zoomperc : float (0-1)
the percentage of the output range to show in the zoomplot, if ‘none’, no zoom plot is added
loc : int
matplotlib.pyplot.legend: location code (0-10)
outputid : int
the output to use whe multiple are compared; starts with 0
Returns: fig : matplotlib.figure.Figure object
figure containing the output
ax1 : axes.AxesSubplot object
the subplot
txtobjects : list of textobjects
enbales the ad hoc replacement of labels when overlapping
Notes
Visualization as proposed by [M2]
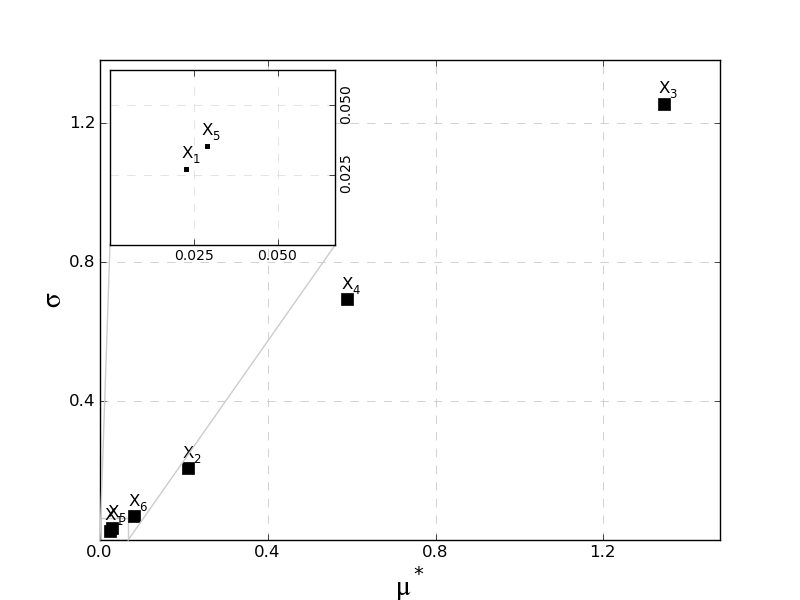
-
MorrisScreening.latexresults(outputid=0, name='Morristable.tex')[source]¶ Print Results in a “deluxetable” Latex
Parameters: outputid : int
teh output to use when evaluation for multiple outputs are calculated
name : str.tex
output file name; use .tex extension in the name
\begin{table}{lccc}
\tablewidth{0pc}
\tablecolumns{4}
\caption{Morris evaluation criteria\label{tab:morris1tot}}
\tablehead{
& $\mu$ & $\mu^*$ & $\sigma$ }
\startdata
$X_1$ & 0.019 & 0.053 & 0.062\\
$X_2$ & -0.014 & 0.28 & 0.37\\
$X_3$ & 0.35 & 1.8 & 2.2\\
$X_4$ & 0.58 & 0.77 & 0.66\\
$X_5$ & -0.0091 & 0.029 & 0.035\\
$X_6$ & 0.023 & 0.088 & 0.11\\
\enddata
\end{table}
-
MorrisScreening.txtresults(outputid=0, name='Morrisresults.txt')[source]¶ Results in txt file to load in eg excel
Parameters: outputid : int
the output to use when evaluation for multiple outputs are calculated
name : str.txt
output file name; use .txt extension in the name
Par mu mustar sigma
$X_1$ 0.01923737 0.05295802 0.06235872
$X_2$ -0.01423402 0.27864841 0.36502709
$X_3$ 0.35127671 1.79863549 2.19419533
$X_4$ 0.57758340 0.76929478 0.66397173
$X_5$ -0.00912314 0.02933937 0.03503700
$X_6$ 0.02319802 0.08834758 0.10503778
Standardized Regression Coefficients (SRC) method¶
Main SRC methods¶
-
class
pystran.SRCSensitivity(parsin, ModelType='external')[source]¶ The regression sensitivity analysis: MC based sampling in combination with a SRC calculation; the rank based approach (less dependent on linearity) is also included in the SRC calculation and is called SRRC
The model is proximated by a linear model of the same parameterspace and the influences of the parameters on the model output is evaluated.
Parameters: parsin : list
Either a list of (min,max,’name’) values, [(min,max,’name’),(min,max,’name’),...(min,max,’name’)] or a list of ModPar instances
Notes
- Rank transformation:
- Rank transformation of the data can be used to transform a nonlinear but monotonic relationship to a linear relationship. When using rank transformation the data is replaced with their corresponding ranks. Ranks are defined by assigning 1 to the smallest value, 2 to the second smallest and so-on, until the largest value has been assigned the rank N.
Examples
>>> ai=np.array([10,20,15,1,2,7]) >>> Xi = [(8.0,9.0,r'$X_1$'),(7.0,10.0,r'$X_2$'),(6.0,11.0,r'$X_3$'), >>> (5.0,12.0,r'$X_4$'), (4.0,13.0,r'$X_5$'),(3.0,4.,r'$X_6$')] >>> #prepare method >>> reg=SRCSensitivity(Xi) >>> #prepare parameter samples >>> reg.PrepareSample(1000) >>> #run model and get outputs for all MC samples >>> output = np.zeros((reg.parset2run.shape[0],1)) >>> for i in range(reg.parset2run.shape[0]): >>> output[i,:] = simplelinear(ai,reg.parset2run[i,:]) >>> #Calc SRC without using rank-based approach >>> reg.Calc_SRC(output, rankbased=False) >>> #check if the sum of the squared values approaches 1 >>> print reg.sumcheck >>> [ 1.00555193] >>> reg.plot_tornado(outputid = 0, ec='grey',fc='grey')
Attributes
_ndim ( intx) number of factors examined. In case the groups are chosen the number of factors is stores in NumFact and sizea becomes the number of created groups, (k) -
Calc_SRC(output, rankbased=False)[source]¶ SRC sensitivity calculation for multiple outputs
Check is done on the Rsq value (higher than 0.7?) and the sum of SRC’s for the usefulness of the method.
Parameters: output : Nxm ndarray
array with the model outputs (N MC simulations and m different outputs)
rankbased : boolean
if True, SRC values are transformed into SRRC values; using ranks instead of values itself
Notes
Least squares Estimation theory, eg. http://www.stat.math.ethz.ch/~geer/bsa199_o.pdf
Attributes
SRC (ndimxnoutputs narray) Standardized Regression Coefficients cova : variance-Covariance matrix core : correlation matrix sumcheck (noutput narray) chek for linearity by summing the SRC values
-
PrepareSample(nbaseruns, samplemethod='Sobol', seedin=1)[source]¶ Sampling of the parameter space. No specific sampling preprocessing procedure is needed, only a general Monte Carlo sampling of the parameter space is expected. To improve the sampling procedure, Latin Hypercube or Sobol pseudo-random sampling can be preferred. The latter is used in the pySTAN framework, but using the ModPar class individually enables Latin Hypercube and random sampling.
Currently only uniform distributions are supported by the framework, but the Modpar class enables other dsitributions to sample the parameter space and bypassing the sampling here.
Parameters: nbaseruns : int
number of samples to take for the analysis; highly dependent from the total number of input factors. range of 10000s is minimum.
samplemethod : ‘sobol’ or ‘lh’
sampling method to use for the sampling
seedin : int
seed to start the Sobol sampling from. This enables to increase the sampling later on, by starting from the previous seedout.
Returns: seed_out : int
the last seed, useful for later use
parset2run : ndarray
parameters to run the model (nbaseruns x ndim)
Notes
- Do the Sobol sampling always for the entire parameter space at the
same time, for LH this doesn’t matter! - Never extend the sampling size with using the same seed, since this generates duplicates of the samples!
Quick analysis of the scatter plots of the ouput versus the parameter values¶
-
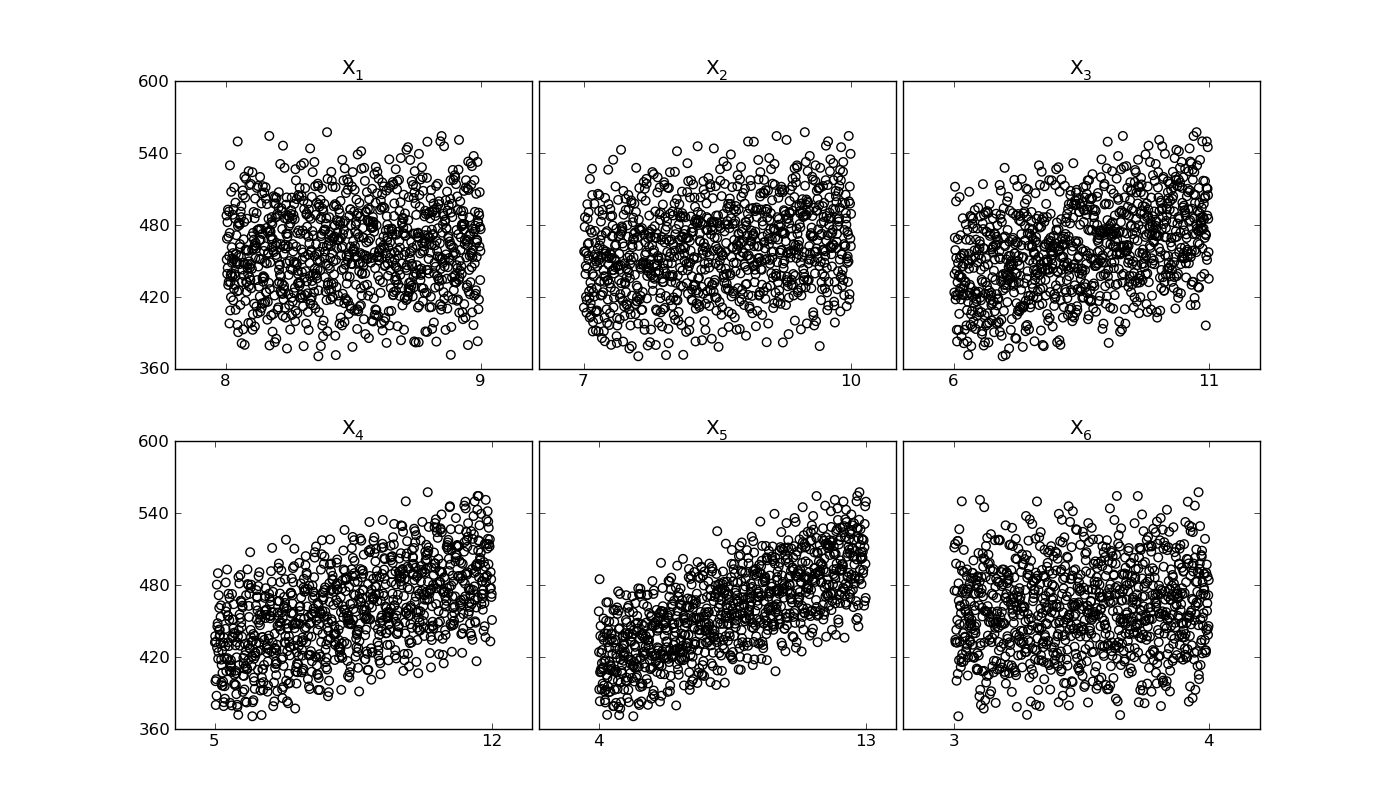
SRCSensitivity.quickscatter(output)[source]¶ Quick link to the general scatter function, by passing to the general scattercheck plot of the sensitivity base-class
Parameters: output: ndimx1 array
array with the output for one output of the model; this can be an Objective function or an other model statistic
Notes
Used to check linearity of the input/output relation in order to evaluate the usefulness of the SRC-regression based technique
Examples
>>> reg.quickscatter(output)
Output image:
Output plots and exports¶
-
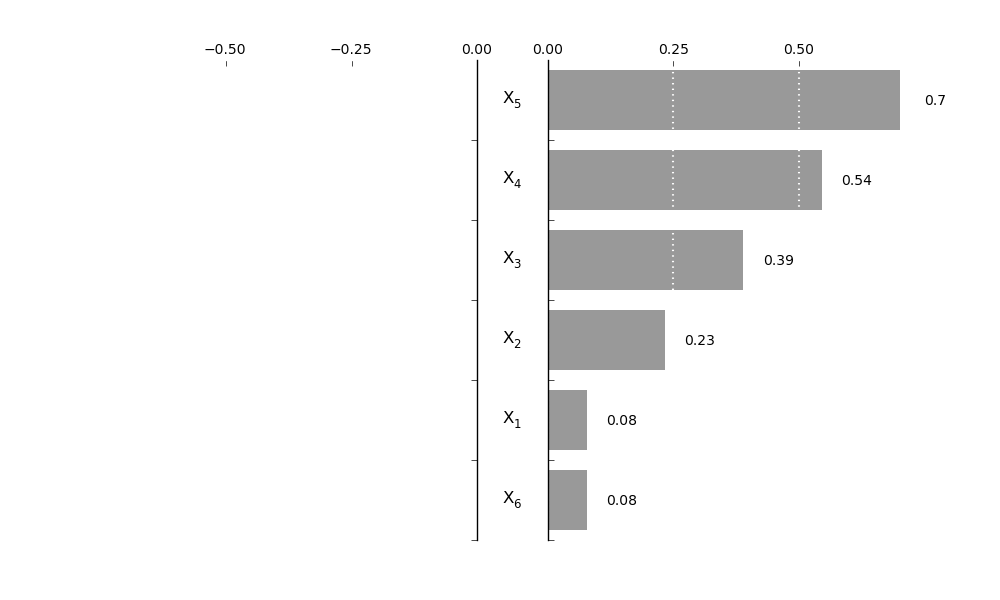
SRCSensitivity.plot_tornado(outputid=0, SRC=True, *args, **kwargs)[source]¶ Make a Tornadplot of the parameter influence on the output; SRC must be calculated first before plotting
Parameters: outputid : int
output for which the tornado plot is made, starting from 0
SRC : boolean
SRC true means that SRC values are used, otherwise the SRRC (ranked!) is used
*args, **kwargs :
arguments passed to the TornadoSensPlot function of the plotfunctions_rev data
Notes
- The parameters for the tornadosensplot are:
- gridbins: int, number of gridlines midwidth: float, width between the two axis, adapt to parameter names setequal: boolean, if True, axis are equal lengths plotnumb: boolean, if True, sensitiviteis are plotted parfontsize: int, font size of the parameter names bandwidth: width of the band
Examples
>>> reg.plot_tornado(outputid = 0,gridbins=3, midwidth=0.2, setequal=True, plotnumb=True, parfontsize=12, bandwidth=0.75)
Output image:
-
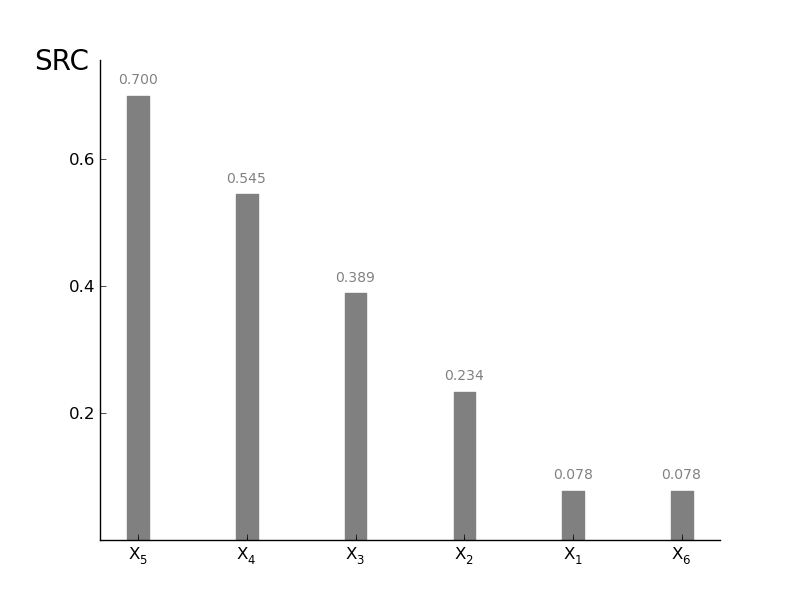
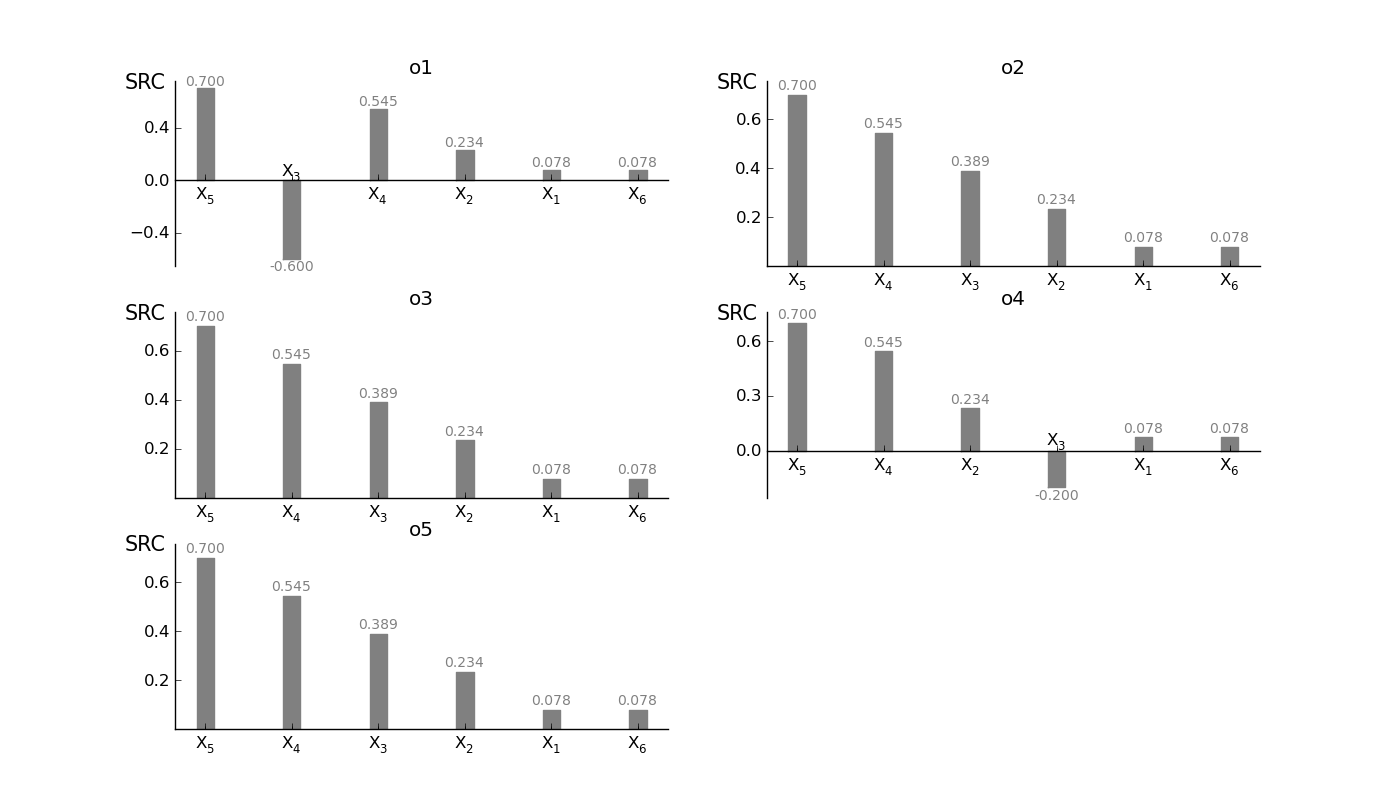
SRCSensitivity.plot_SRC(width=0.2, addval=True, sortit=True, outputid='all', ncols=3, outputnames=[], *args, **kwargs)[source]¶ Plot a barchart of the SRC values; actually a Tornadoplot in the horizontal direction. More in the style of the other methods.
TODO: make for subset of outputs, also in other methods; currently all or 1
Parameters: width : float (0-1)
width of the bars in the barchart
addval : bool
if True, the sensitivity values are added to the graph
sortit : bool
if True, larger values (in absolute value) are plotted closest to the y-axis
outputid : int or all
the output to use whe multiple are compared; starts with 0 if all, the different outputs are plotted in subplots
ncols : int
number of columns to put the subplots in
outputnames : list of strings
[] to plot no outputnames, otherwise list of strings equal to the number of outputs
*args, **kwargs : args
passed to the matplotlib.bar
Returns: fig : matplotlib.figure.Figure object
figure containing the output
ax1 : axes.AxesSubplot object
the subplot
Examples
>>> #plot single SRC >>> reg.plot_SRC(outputid=0, width=0.3, sortit = True, ec='grey',fc='grey') >>> #plot single SRC (assume 4 outputs) >>> reg.plot_SRC(outputid = 'all' , ncols = 2, outputnames=['o1','o2','o3','o4'], ec='grey', fc='grey')
Output image if a single output is selected:
Output image if a all outputs are selected:
-
SRCSensitivity.latexresults(outputnames, rank=False, name='SRCtable.tex')[source]¶ Print results SRC values or ranks in a “deluxetable” Latex
Parameters: outputnames : list of strings
The names of the outputs in the colomns
rank : boolean
if rank is True, rankings or plotted in tabel instead of the SRC values
name : str.tex
output file name; use .tex extension in the name
\begin{table}{lccccc}
\tablewidth{0pc}
\tablecolumns{6}
\caption{Global SRC parameter ranking\label{tab:SRCresult}}
\tablehead{
& o1 & o2 & o3 & o4 & o5 }
\startdata
$X_1$ & 5 & 5 & 5 & 5 & 5\\
$X_2$ & 4 & 4 & 4 & 4 & 4\\
$X_3$ & 3 & 3 & 3 & 3 & 3\\
$X_4$ & 2 & 2 & 2 & 2 & 2\\
$X_5$ & 1 & 1 & 1 & 1 & 1\\
$X_6$ & 6 & 6 & 6 & 6 & 6\\
\enddata
\end{table}
-
SRCSensitivity.txtresults(outputnames, rank=False, name='SRCresults.txt')[source]¶ Results rankmatrix in txt file to load in eg excel
Parameters: outputnames : list of strings
The names of the outputs in the colomns
rank : boolean
if rank is True, rankings or plotted in tabel instead of the SRC values
name : str.txt
output file name; use .txt extension in the name
Par o1 o2 o3 o4 o5
$X_1$ 0.0778505546741 0.0778505546741 0.0778505546741 0.0778505546741 0.0778505546741
$X_2$ 0.233670804823 0.233670804823 0.233670804823 0.233670804823 0.233670804823
$X_3$ -0.6 0.389115599046 0.389115599046 0.389115599046 0.389115599046
$X_4$ 0.544743494911 0.544743494911 0.544743494911 0.544743494911 0.544743494911
$X_5$ 0.700482475678 0.700482475678 0.700482475678 0.700482475678 0.700482475678
$X_6$ 0.0778270555871 0.0778270555871 0.0778270555871 0.0778270555871 0.0778270555871
Sobol Variance based method¶
Main Sobol variance methods¶
-
class
pystran.SobolVariance(parsin, ModelType='pyFUSE')[source]¶ Sobol Sensitivity Analysis Variance Based
Parameters: ParsIn : list
either a list of (min,max,’name’) values, [(min,max,’name’),(min,max,’name’),...(min,max,’name’)] or a list of ModPar instances
ModelType : pyFUSE | PCRaster | external
Give the type of model working with
Notes
Calculates first and total order, and second order Total Sensitivity, according to [S1] , higher order terms and bootstrapping is not (yet) included
References
[S1] (1, 2, 3, 4) Saltelli, A., P. Annoni, I. Azzini, F. Campolongo, M. Ratto, S. Tarantola,(2010) Variance based sensitivity analysis of model output. Design and estimator for the total sensitivity index, Computer Physics Communications, 181, 259–270 [S2] (1, 2) Saltelli, Andrea, Marco Ratto, Terry Andres, Francesca Campolongo, Jessica Cariboni, Debora Gatelli, Michaela Saisana, and Stefano Tarantola. Global Sensitivity Analysis, The Primer. John Wiley & Sons Ltd, 2008. Examples
>>> ai=[0, 1, 4.5, 9, 99, 99, 99, 99] >>> alphai = 0.5 * np.ones(len(ai)) >>> Xi = [(0.0,1.0,'par1'),(0.0,1.0,'par2'),(0.0,1.0,'par3'),(0.0,1.0,'par4'), (0.0,1.0,'par5'),(0.0,1.0,'par6'),(0.0,1.0,'par7'),(0.0,1.0,'par8'),] >>> di=np.random.random(8) >>> #set up sensitivity analysis >>> sens2 = SobolVariance(Xi, ModelType = 'testmodel') >>> #Sampling strategy >>> sens2.SobolVariancePre(1500) >>> #Run model + evaluate the output >>> sens2.runTestModel('analgfunc', [ai, alphai, di]) #here normally other model >>> #results in txt file >>> sens2.txtresults(name='SobolTestOut.txt') >>> #results in tex-table >>> sens2.latexresults(name='SobolTestOut.tex') >>> #Plots >>> sens2.plotSi(ec='grey',fc='grey') >>> sens2.plotSTi(ec='grey',fc='grey') >>> sens2.plotSTij() >>> Si_evol, STi_evol = sens2.sens_evolution(color='k')
Attributes
parset2run (ndarray) every row is a parameter set to run the model for. All sensitivity methods have this attribute to interact with base-class running -
SobolVariancePost(output, repl=1, adaptedbaserun=None, forevol=False)[source]¶ Calculate first and total indices based on model output and sampled values
the sensmatrices for replica, have repititions in the rows, columns are the factors
only output => make multiple outputs (todo!!), by using the return, different outputs can be tested...
Parameters: output : ndarray
Nx1 matrix with the model outputs
repl : int
number of replicates used
adaptedbaserun : int
number of baseruns to base calculations on
forevol : boolean
True if used for evaluating the evolution
Notes
The calculation methods follows as the directions given in [S1]
-
SobolVariancePre(nbaseruns, seed=1, repl=1, conditional=None)[source]¶ Set up the sampling procedure of N*(k+2) samples
Parameters: nbaseruns : int
number of samples for the basic analysis, total number of model runs is Nmc(k+2), with k the number of factors
seed : int
to set the seed point for the sobol sampling; enables to enlarge a current sample
repl : int
Replicates the entire sampling procedure. Can be usefull to test if the current sampling size is large enough to get convergence in the sensitivity results (bootstrapping is currently not included)
Notes
Following the Global sensitivity analysis, [S2], but updated for [S1] for more optimal convergence of the sensitivity indices
-
Sobolsampling(nbaseruns, seedin=1)[source]¶ Performs Sobol sampling procedure, given a set of ModPar instances, or by a set of (min,max) values in a list.
This function is mainly used as help function, but can be used to perform Sobol sampling for other purposes
Todo: improve seed handling, cfr. bioinspyred package
Parameters: nbaseruns : int
number of runs to do
seed : int
to et the seed point for the sobol sampling
Notes
DO SOBOL SAMPLING ALWAYS FOR ALL PARAMETERS AT THE SAME TIME!
Duplicates the entire parameter set to be able to divide in A and B matric
Examples
>>> p1 = ModPar('tp1',2.0,4.0,3.0,'randomUniform') >>> p2 = ModPar('tp2',1.0,5.0,4.0,'randomUniform') >>> p3 = ModPar('tp3',0.0,1.0,0.1,'randomUniform') >>> sobanal1 = SobolVariance([p1,p2,p3]) >>> sobanal2 = SobolVariance([(2.0,4.0,'tp1'),(1.0,5.0,'tp2'),(0.0,1.0,'tp3')]) >>> pars1 = sobanal.Sobolsampling(100,seed=1) >>> pars2 = sobanal.Sobolsampling(100,seed=1)
-
runTestModel(model, inputsmod, repl=1)[source]¶ Run a testmodel
Use a testmodel to get familiar with the method and try things out. The implemented methods are * G Sobol function: testfunction with analytical solution * gstarfunction: testfunction with analytical solution, extended version of the G sobol function
Parameters: model : string
name fo the testmodel
inputsmod : list
list with all the inputs of the model, except of the sampled stuff
repl : int
Notes
-
Check the convergence of the analysis¶
-
SobolVariance.sens_evolution(output=None, repl=1, labell=-0.07, *args, **kwargs)[source]¶ Check the convergence of the current sequence
Parameters: output : optional
if True; this output is used, elsewhere the generated output
repl : int
must be 1 to work
labell : float
aligns the ylabels
*args, **kwargs : args
passed to the matplotlib.plot function
Returns: Si_evol : ndarray
Si of the factors in number of nbaseruns
STi_evol : ndarray
STi of the factors in number of nbaseruns
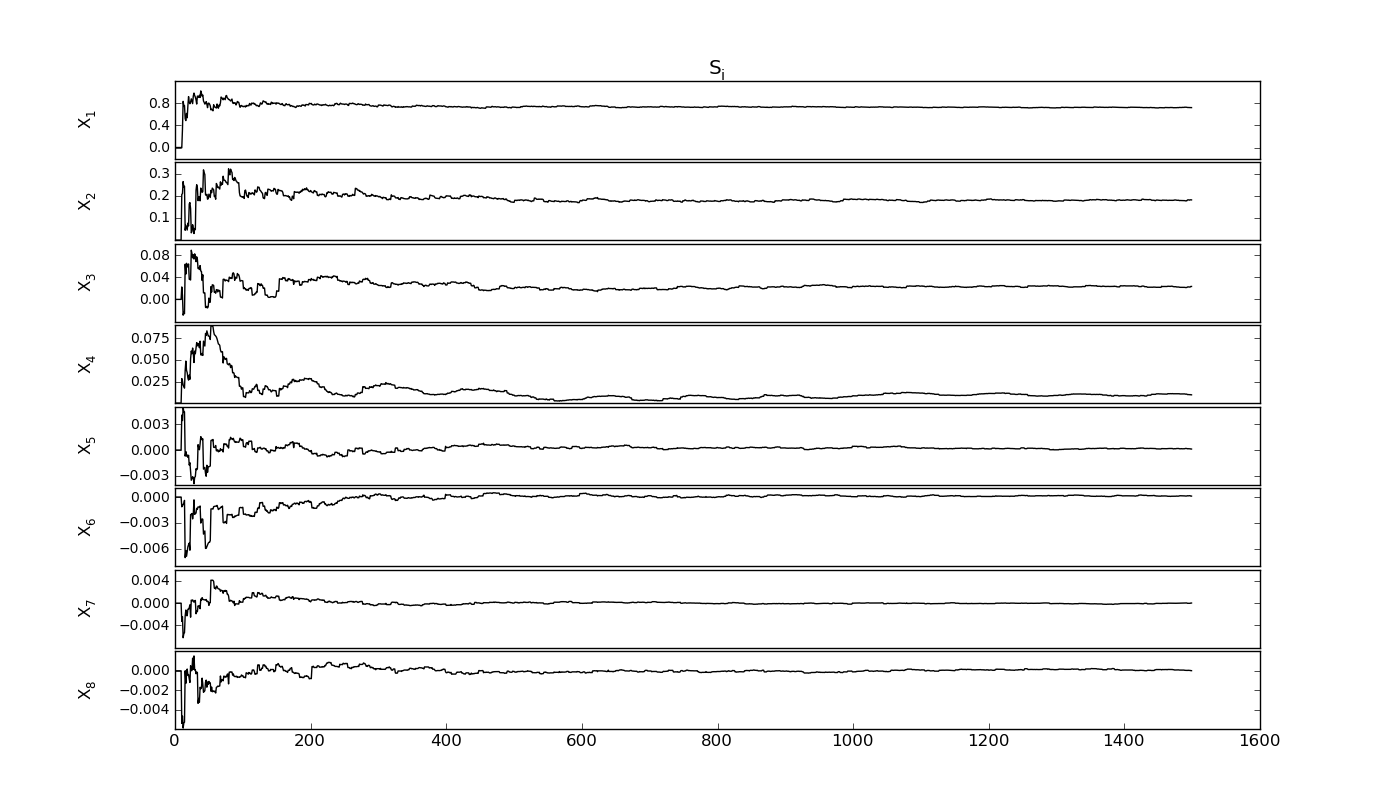
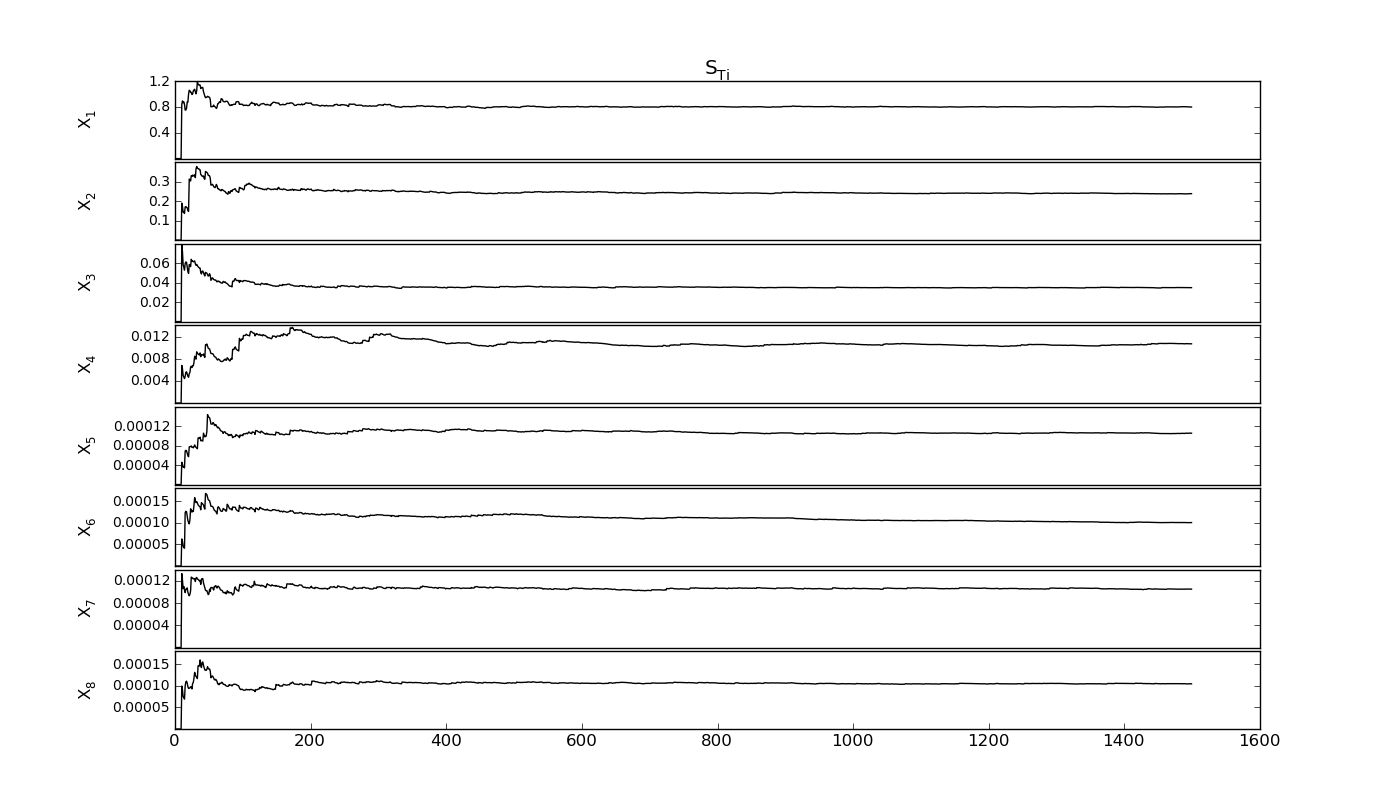
Output plots and exports¶
-
SobolVariance.plotSTij(linewidth=2.0)[source]¶ Plot the STij interactive terms
Parameters: linewidth : float
width of the black lines of the grid
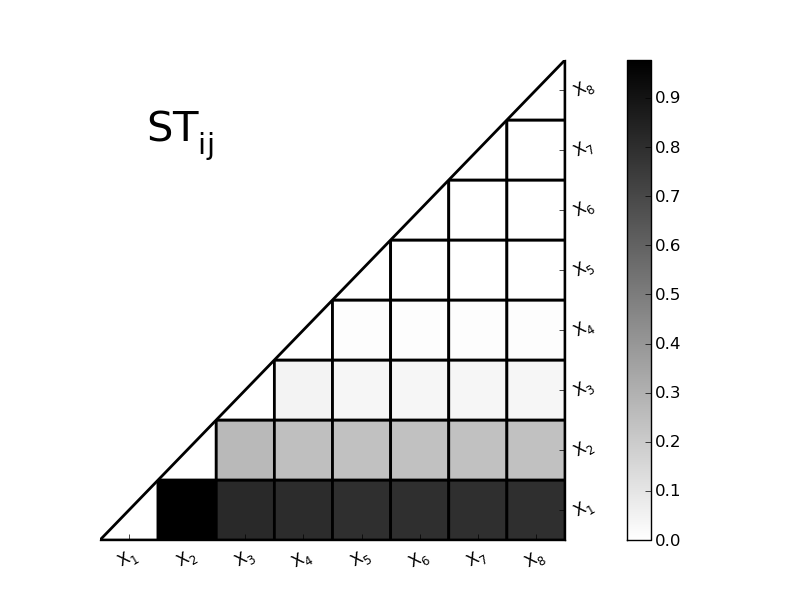
-
SobolVariance.plotSTi(width=0.5, addval=True, sortit=True, *args, **kwargs)[source]¶ Plot a barchart of the given STi
Parameters: width : float (0-1)
width of the bars in the barchart
addval : bool
if True, the morris mu values are added to the graph
sortit : bool
if True, larger values (in absolute value) are plotted closest to the y-axis
*args, **kwargs : args
passed to the matplotlib.bar
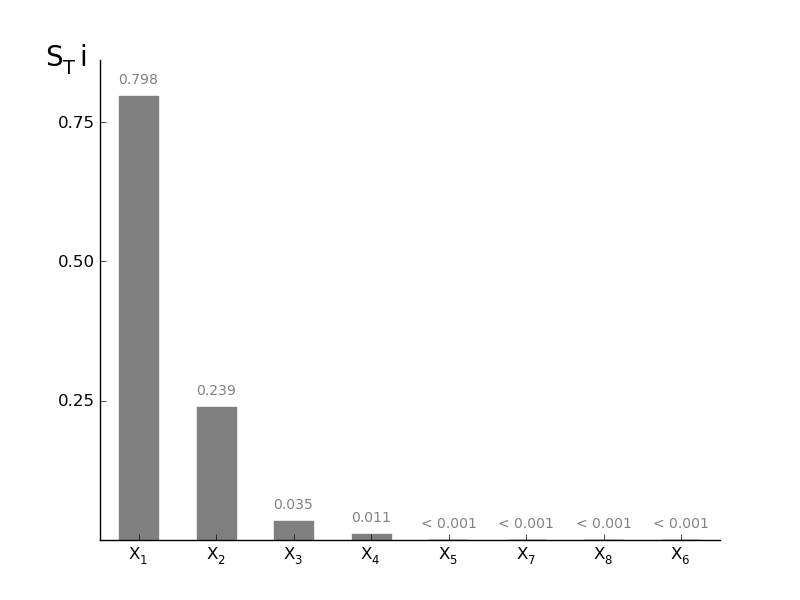
-
SobolVariance.plotSi(width=0.5, addval=True, sortit=True, *args, **kwargs)[source]¶ Plot a barchart of the given Si
Parameters: width : float (0-1)
width of the bars in the barchart
addval : bool
if True, the morris mu values are added to the graph
sortit : bool
if True, larger values (in absolute value) are plotted closest to the y-axis
*args, **kwargs : args
passed to the matplotlib.bar
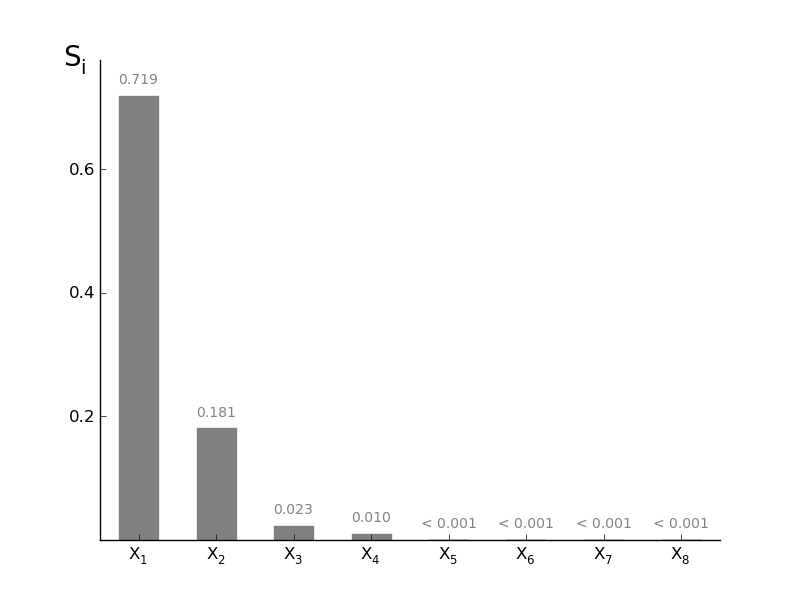
-
SobolVariance.latexresults(name='Soboltable.tex')[source]¶ Print Results in a “deluxetable” Latex
Parameters: name : str.tex
output file name; use .tex extension in the name
\begin{table}{lcc}
\tablewidth{0pc}
\tablecolumns{3}
\caption{First order and Total sensitivity index\label{tab:sobol1tot}}
\tablehead{
& $S_i$ & $S_{Ti}$ }
\startdata
par1 & 0.72 & 0.79\\
par2 & 0.18 & 0.24\\
par3 & 0.023 & 0.034\\
par4 & 0.0072 & 0.010\\
par5 & 0.000044 & 0.00011\\
par6 & 0.000068 & 0.00011\\
par7 & 0.000028 & 0.00010\\
par8 & 0.000049 & 0.00011\\
SUM & 0.93 & 1.1\\
\enddata
\end{table}
-
SobolVariance.txtresults(name='Sobolresults.txt')[source]¶ Results in txt file to load in eg excel
Parameters: name : str.txt
output file name; use .txt extension in the name
Par Si STi
par1 0.71602494 0.78797717
par2 0.17916175 0.24331297
par3 0.02346677 0.03437614
par4 0.00720168 0.01044238
par5 0.00004373 0.00010526
par6 0.00006803 0.00010556
par7 0.00002847 0.00010479
par8 0.00004918 0.00010522
SUM 0.92604455 1.07652948
Latin-Hypercube OAT method¶
Main GLobal OAT methods¶
-
class
pystran.GlobalOATSensitivity(parsin, ModelType='external')[source]¶ A merged apporach of sensitivity analysis; also extended version of the LH-OAT approach
Parameters: parsin : list
either a list of (min,max,’name’) values, [(min,max,’name’),(min,max,’name’),...(min,max,’name’)] or a list of ModPar instances
Notes
Original method described in [OAT1], but here generalised in the framework, by adding different measures of sensitivity making the sampling method adjustable (lh or Sobol pseudosampling) and adding the choice of numerical approach to derive the derivative.
References
[OAT1] (1, 2) van Griensven, Ann, T. Meixner, S. Grunwald, T. Bishop, M. Diluzio, and R. Srinivasan. ‘A Global Sensitivity Analysis Tool for the Parameters of Multi-variable Catchment Models’ 324 (2006): 10–23. [OAT2] (1, 2) Pauw, Dirk D E, D E Leenheer Decaan, and V A N Langenhove Promotor. OPTIMAL EXPERIMENTAL DESIGN FOR CALIBRATION OF BIOPROCESS MODELS : A VALIDATED SOFTWARE TOOLBOX OPTIMAAL EXPERIMENTEEL ONTWERP VOOR KALIBRERING VAN BIOPROCESMODELLEN : EEN GEVALIDEERDE SOFTWARE TOOLBOX BIOPROCESS MODELS : A VALIDATED SOFTWARE TOOLBOX, 2005. Examples
>>> #EXAMPLE WITH ANALG-FUNCTION >>> #variables and parameters >>> ai=np.array([78, 12, 0.5, 2, 97, 33]) >>> Xi = [(0.0,1.0,r'$X_1$'),(0.0,1.0,r'$X_2$'),(0.0,1.0,r'$X_3$'), (0.0,1.0,r'$X_4$'), (0.0,1.0,r'$X_5$'),(0.,1,r'$X_6$')] >>> #prepare model class >>> goat=GlobalOATSensitivity(Xi) >>> #prepare the parameter sample >>> goat.PrepareSample(100,0.01, samplemethod='lh', numerical_approach='central') >>> #calculate model outputs >>> output = np.zeros((goat.parset2run.shape[0],1)) >>> for i in range(goat.parset2run.shape[0]): output[i,:] = analgfunc(ai,goat.parset2run[i,:]) >>> #evaluate sensitivitu >>> goat.Calc_sensitivity(output) >>> #transform sensitivity into ranking >>> goat.Get_ranking() >>> #plot the partial effect based sensitivity >>> goat.plotsens(indice='PE', ec='grey',fc='grey') >>> #plot the rankmatrix >>> goat.plot_rankmatrix(outputnames)
Attributes
_ndim ( intx) number of factors examined. In case the groups are chosen the number of factors is stores in NumFact and sizea becomes the number of created groups, (k) -
Calc_sensitivity(output)[source]¶ Calculate the Central or Single Absolute Sensitvity (CAS), the Central or Single Total Sensitivity (CTRS) and the Partial Effect (PE) of the different outputs given. These outputs can be either a set of model objective functions or a timerserie output
Parameters: output: Nxndim array
array with the outputs for the different outputs of the model; this can be an Objective function, or a timeserie of the model output
Notes
PE is described in [OAT1], the other two are just global extensions of the numercial approach to get local sensitivity results, see [OAT2].
Attributes
CAS_SENS: Central/Single Absolute Sensitivity CTRS_SENS: Central/Single Total Sensitivity PE_SENS: Partial effect (van Griensven)
-
PrepareSample(nbaseruns, perturbation_factor, samplemethod='Sobol', numerical_approach='central', seedin=1)[source]¶ Sampling of the parameter space. No specific sampling preprocessing procedure is needed, only a general Monte Carlo sampling of the parameter space is expected. To improve the sampling procedure, Latin Hypercube or Sobol pseudo-random sampling can be preferred. The latter is used in the pySTAN framework, but using the ModPar class individually enables Latin Hypercube and random sampling.
Currently only uniform distributions are supported by the framework, but the Modpar class enables other dsitributions to sample the parameter space and bypassing the sampling here.
Parameters: nbaseruns : int
number of samples to take for the analysis; highly dependent from the total number of input factors. range of 10000s is minimum.
perturbation_factor : float or list
if only one number, all parameters get the same perturbation factor otherwise, _ndim elements in list
samplemethod : ‘sobol’ or ‘lh’
sampling method to use for the sampling
numerical_approach : ‘central’ or ‘single’
central approach needs n(2*k) runs, singel only n(k+1) runs; OAT calcluation depends on this.
seedin : int
seed to start the Sobol sampling from. This enables to increase the sampling later on, by starting from the previous seedout.
Returns: seed_out : int
the last seed, useful for later use
parset2run : ndarray
parameters to run the model (nbaseruns x ndim)
Notes
- Do the Sobol sampling always for the entire parameter space at the
same time, for LH this doesn’t matter * Never extend the sampling size with using the same seed, since this generates duplicates of the samples * More information about the central or single numerical choice is given in [OAT2].
-
Output plots and exports¶
-
GlobalOATSensitivity.Get_ranking(choose_output=False)[source]¶ Only possible if Calc_sensitivity is already finished; every columns gets is one ranking and the overall_importance is calculated based on the lowest value of the other rankings (cfr. Griensven)
rankmatrix: defines the rank of the parameter self.rankdict: defines overall rank of the parameters with name
Parameters: choose_output: integer
starting from 1 as the first output
Notes
TODO make a dataframe of pandas as output: rows is par, cols is output
Attributes
rankdict (only when single output selected): Dictionary giving the ranking of the parameter rankmatrix: Main output: gives for each parameter (rows) the ranking for the different outputs overall_importance: Returns the summarized importance of the parameter over the different outputs, by checking the minimal ranking of the parameters
-
GlobalOATSensitivity.plotsens(indice='PE', width=0.5, addval=True, sortit=True, outputid=0, *args, **kwargs)[source]¶ Plot a barchart of either CAS, CTRS or PE based Senstivitity
Parameters: indice: PE, CAS or CTRS
choose which indice you want to show
width : float (0-1)
width of the bars in the barchart
addval : bool
if True, the sensitivity values are added to the graph
sortit : bool
if True, larger values (in absolute value) are plotted closest to the y-axis
outputid : int
the output to use whe multiple are compared; starts with 0
*args, **kwargs : args
passed to the matplotlib.bar
Returns: fig : matplotlib.figure.Figure object
figure containing the output
ax1 : axes.AxesSubplot object
the subplot
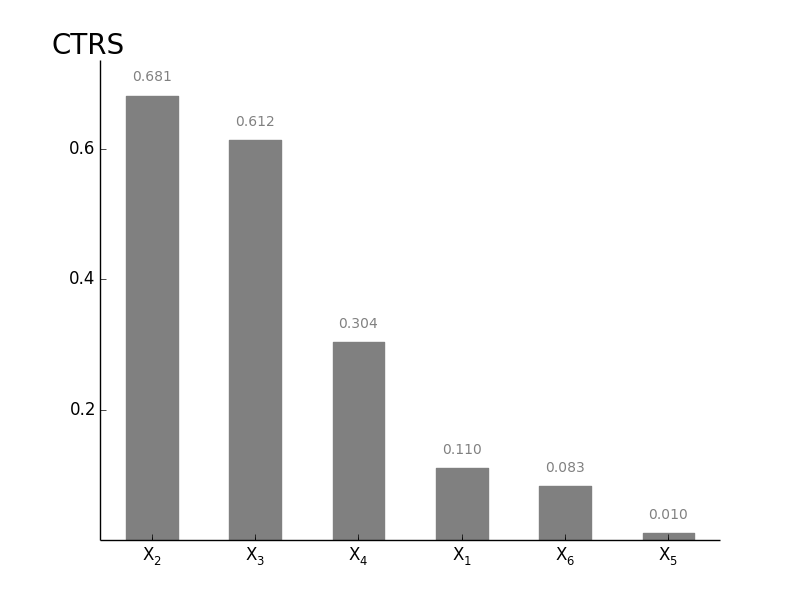
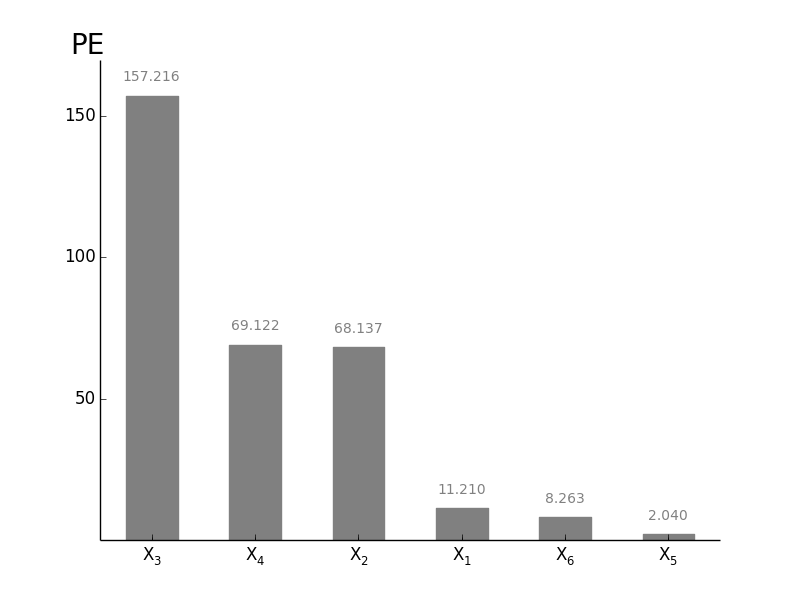
-
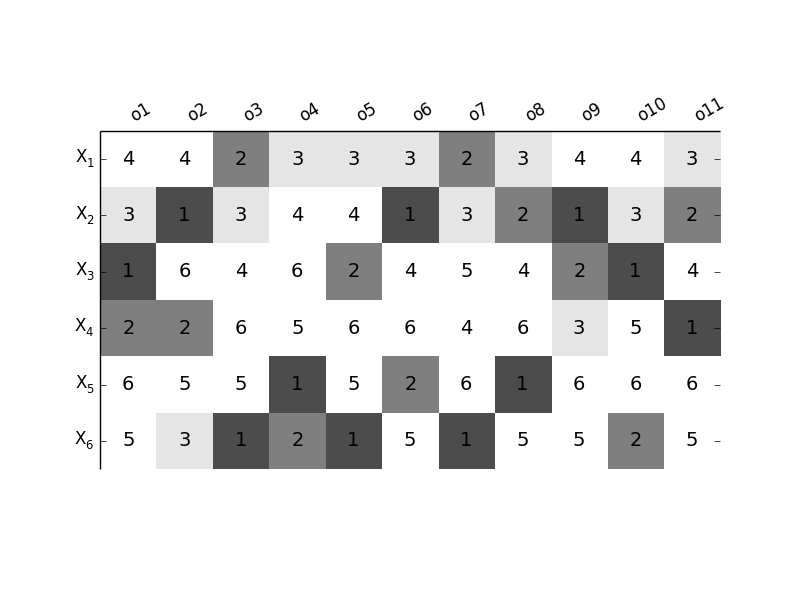
GlobalOATSensitivity.plot_rankmatrix(outputnames, fontsize=14)[source]¶ Make an overview plot of the resulting ranking of the different parameters for the different outputs
Parameters: outputnames : list of strings
The names of the outputs in the colomns
fontsize: int
size of the numbers of the ranking
-
GlobalOATSensitivity.latexresults(outputnames, name='GlobalOATtable.tex')[source]¶ Print results rankmatrix in a “deluxetable” Latex
Parameters: outputnames : list of strings
The names of the outputs in the colomns
name : str.tex
output file name; use .tex extension in the name
\begin{table}{lccccccccccc}
\tablewidth{0pc}
\tablecolumns{12}
\caption{Global OAT parameter ranking\label{tab:globaloatrank}}
\tablehead{
& o1 & o2 & o3 & o4 & o5 & o6 & o7 & o8 & o9 & o10 & o11 }
\startdata
$X_1$ & 4 & 4 & 2 & 3 & 3 & 3 & 2 & 3 & 4 & 4 & 3\\
$X_2$ & 3 & 1 & 3 & 4 & 4 & 1 & 3 & 2 & 1 & 3 & 2\\
$X_3$ & 1 & 6 & 4 & 6 & 2 & 4 & 5 & 4 & 2 & 1 & 4\\
$X_4$ & 2 & 2 & 6 & 5 & 6 & 6 & 4 & 6 & 3 & 5 & 1\\
$X_5$ & 6 & 5 & 5 & 1 & 5 & 2 & 6 & 1 & 6 & 6 & 6\\
$X_6$ & 5 & 3 & 1 & 2 & 1 & 5 & 1 & 5 & 5 & 2 & 5\\
\enddata
\end{table}
-
GlobalOATSensitivity.txtresults(outputnames, name='GlobalOATresults.txt')[source]¶ Results rankmatrix in txt file to load in eg excel
Parameters: outputnames : list of strings
The names of the outputs in the colomns
name : str.txt
output file name; use .txt extension in the name
Par o1 o2 o3 o4 o5 o6 o7 o8 o9 o10 o11
$X_1$ 4 4 2 3 3 3 2 3 4 4 3
$X_2$ 3 1 3 4 4 1 3 2 1 3 2
$X_3$ 1 6 4 6 2 4 5 4 2 1 4
$X_4$ 2 2 6 5 6 6 4 6 3 5 1
$X_5$ 6 5 5 1 5 2 6 1 6 6 6
$X_6$ 5 3 1 2 1 5 1 5 5 2 5
Dynamic Identifiability Analysis (DYNIA)¶
added soon
Regional Sensitivity Analysis (RSA)¶
Extra info added soon
Main RSA methods¶
-
class
pystran.RegionalSensitivity(parsin, ModelType='pyFUSE')[source]¶ Regional Sensitivity Analysis (Monte Carlo Filtering).
Parameters: ParsIn : list
either a list of (min,max,’name’) values, [(min,max,’name’),(min,max,’name’),...(min,max,’name’)] or a list of ModPar instances
ModelType : pyFUSE | PCRaster | external
Give the type of model working with
Notes
The method basically ranks/selects parameter sets based on a evaluation criterion and checks the marginal influence of the different parameters on this criterion. In literature, the method is known as Regional Sensitivity Analysis (RSA, [R1]), but also describe in [R2] and referred to as Monte Carlo Filtering. Intially introduced by [R1] with a split between behavioural and non-behavioural accombined with a Kolmogorov- smirnov rank test (necessary, but nof sufficient to determine insensitive), applied with ten-bins split of the behavioural by [R3] and a ten bins split of the entire parameter range by [R4].
The method is directly connected to the GLUE approach, using the behavioural simulation to define prediction limits of the model output. Therefore, this class is used as baseclass for the GLUE uncertainty.
References
[R1] (1, 2) Hornberger, G.M., and R.C. Spear. An Approach to the Preliminary Analysis of Environmental Systems. Journal of Environmental Management 12 (1981): 7–18. [R2] Saltelli, Andrea, Marco Ratto, Terry Andres, Francesca Campolongo, Jessica Cariboni, Debora Gatelli, Michaela Saisana, and Stefano Tarantola. Global Sensitivity Analysis, The Primer. John Wiley & Sons Ltd, 2008. [R3] Freer, Jim, Keith Beven, and Bruno Ambroise. Bayesian Estimation of Uncertainty in Runoff Prediction and the Value of Data: An Application of the GLUE Approach. Water Resources Research 32, no. 7 (1996): 2161. http://www.agu.org/pubs/crossref/1996/95WR03723.shtml. [R4] Wagener, Thorsten, D. P. Boyle, M. J. Lees, H. S. Wheater, H. V. Gupta, and S. Sorooshian. A Framework for Development and Application of Hydrological Models. Hydrology and Earth System Sciences 5, no. 1 (2001): 13–26. -
PrepareSample(nbaseruns, seedin=1)[source]¶ Sampling of the parameter space. No specific sampling preprocessing procedure is needed, only a general Monte Carlo sampling of the parameter space is expected. To improve the sampling procedure, Latin Hypercube or Sobol pseudo-random sampling can be preferred. The latter is used in the pySTAN framework, but using the ModPar class individually enables Latin Hypercube and random sampling.
Currently only uniform distributions are supported by the framework, but the Modpar class enables other dsitributions to sample the parameter space and bypassing the sampling here.
Parameters: nbaseruns : int
number of samples to take for the analysis; highly dependent from the total number of input factors. range of 10000s is minimum.
seedin : int
seed to start the Sobol sampling from. This enables to increase the sampling later on, by starting from the previous seedout.
Returns: seed_out : int
the last seed, useful for later use
parset2run : ndarray
parameters to run the model (nbaseruns x ndim)
Notes
- Do the Sobol sampling always for the entire parameter space at the
same time, for LH this doesn’t matter * Never extend the sampling size with using the same seed, since this generates duplicates of the samples
-
select_behavioural(output, method='treshold', threshold=0.0, percBest=0.2, norm=True, mask=True)[source]¶ Select bevahvioural parameter sets, based on output evaluation criterion used. w The algorithm makes only a ‘mask’ for further operation, in order to avoid memory overload by copying matrices
Parameters: threshold :
output : ndarray
Nx1
Method can be ‘treshold’ based or ‘percentage’ based
All arrays must have same dimension in 0-direction (vertical); output-function only 1 column
InputPar is nxnpar; output is nx1; Output nxnTimesteps
this most be done for each structure independently
Output of OFfunctions/Likelihoods normalised or not (do it if different likelihoods have to be plotted together)
-
Generalised Likelihood Uncertainty Estimation (GLUE)¶
added soon